1 | Add to Reading ListSource URL: www.borealgenomics.comLanguage: English - Date: 2012-10-12 18:57:42
|
|---|
2 | Add to Reading ListSource URL: www.sagescience.comLanguage: English - Date: 2014-04-01 10:06:01
|
|---|
3 | Add to Reading ListSource URL: www.abnova.comLanguage: English - Date: 2008-12-29 06:50:18
|
|---|
4 | Add to Reading ListSource URL: www.abnova.comLanguage: English - Date: 2008-12-29 06:38:49
|
|---|
5 | Add to Reading ListSource URL: www.abnova.comLanguage: English - Date: 2008-12-29 06:39:36
|
|---|
6 | Add to Reading ListSource URL: www.cilr.uq.edu.auLanguage: English - Date: 2011-09-29 19:37:49
|
|---|
7 | Add to Reading ListSource URL: www.borealgenomics.comLanguage: English - Date: 2013-01-24 18:27:18
|
|---|
8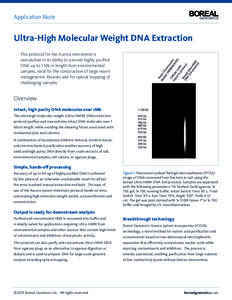 | Add to Reading ListSource URL: www.borealgenomics.comLanguage: English - Date: 2013-01-24 18:27:19
|
|---|
9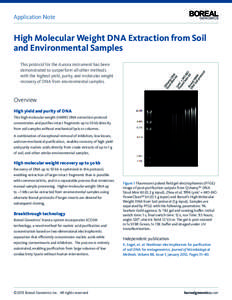 | Add to Reading ListSource URL: www.borealgenomics.comLanguage: English - Date: 2013-01-24 18:27:17
|
|---|
10 | Add to Reading ListSource URL: www.ncbe.reading.ac.ukLanguage: English - Date: 2000-05-23 13:41:17
|
|---|